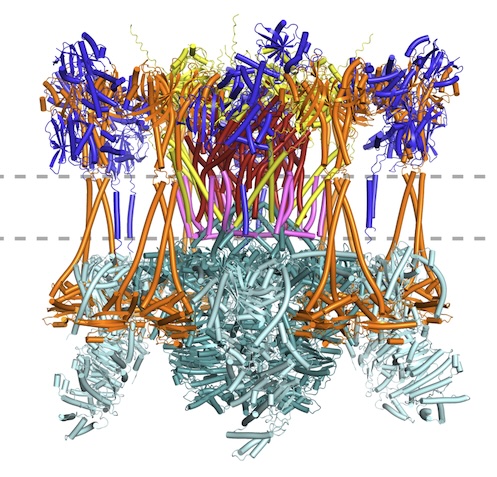
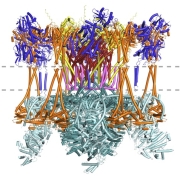
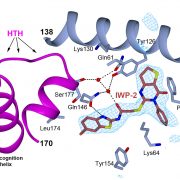
New article: Advances in protein structure prediction highlight unexpected commonalities between Gram-positive and Gram-negative conjugative T4SS
New article from the Berntsson lab, where we have identified which components are conserved across all conjugative Type 4 Secretion Systems, and used the new modelling tools available to model the pCF10 T4SS structure. Published now in JMB:
https://www.sciencedirect.com/science/article/pii/S0022283624005540?via%3Dihub