The Wolf-Watz NMR lab
In my lab we are interested in mechanistic aspects of protein function with a special focus on enzyme dynamics. We are using state of the art biophysical and structural biology tools to probe the relationships between structure, function and dynamics. Our main tool is solution state NMR spectroscopy and x-ray crystallography and supplemented with computational structural biology. Our focus areas are fundamental enzyme function, human protein kinases linked to cancer, infection biology and biotechnology.
News flash: February 8 2023. We just got a paper accepted in “Journal of Chemical Information and Modeling”
News flash: February 4 2023. Our perspective article is accepted in “Journal of Structural Dynamics.
News flash: February 6 2023. Léon Jona Schierholz joined the lab as a PhD student
News flash: February 2023. Tamás Milan Nagy joined the lab as a postdoc.
News flash: October 2022. Jhonalex Buitrago joined the lab as a postdoc.
News flash: October 2022. Apoorvs paper was accepted in “Science Advances”
 Lab picture @January 2022
Lab picture @January 2022
DISCOVERIES IN PROTEIN SCIENCE
Rational design of enzyme catalyzing novel chemical reactions is one of the outstanding goals in contemporary protein science that, so far, only has been partially realized and then mostly for “simple” elimination reactions. Therefore, and in order to reach this goal experimental approaches that can uncover new and fundamental principles in enzymatic catalysis, including understanding of both binding and the chemical step are needed. In my lab we are utilizing evolutionary “time-travel” both backwards and forward in time for fundamental discoveries observed through a lens of biophysical and structural studies. For backwards looking we are exploiting the discovery of the Asgard archaea that are believed to be the closest known relatives to eukaryotic cells. From a comparative approach with Adenylate kinases isolated from Odinarchaeota, E.coli and human we have uncovered the molecular underpinning of enzymatic multi-selectivity and an unexpected oligomerisation dependency of an active site. Further, we have uncovered new insight into the role of magnesium ions in catalysis of the adenylate kinase reaction (ATP + AMP <-> 2ADP). To read more please visit: www.science.org/doi/10.1126/sciadv.abm4089.
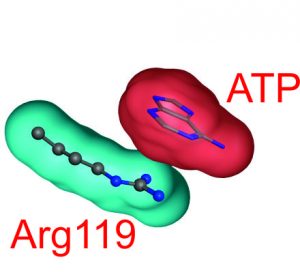
We have discovered a key element in activation of adenylate kinase. Nucleation of the activating conformational change of adenylate kinase occurs via a strong cation-π interaction formed between the cationic sidechain of an arginine and the aromatic adenosine base of ATP. The discovery may pave way for future enzyme design efforts aimed at recognition of aromatic substrates. A multidisciplinary effort (NMR, XRD, DFT calculations) enabled the finding that is published in biochemistry. https://pubs.acs.org/doi/10.1021/acs.biochem.9b00538
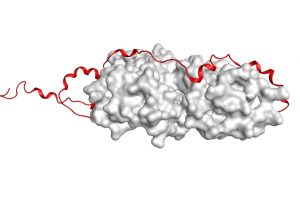
We have discovered a new mechanism for protein-protein interactions that we denote “coupled disordering and binding”, For details read our 2018 JMB paper: “Formation of a translocation competent protein complex by a dynamic wrap-around binding mechanism“. Gupta et al, JMB, 2018,430, 3157-3169
 See our 2018 paper in PNAS: “Molecular Mechanism of ATP versus GTP selectivity of adenylate kinase” The article is based on nice combo of NMR and X-ray!!! (Rogne, et al, PNAS, 2018, 115, 3012-3017 6F7U.pdb).
See our 2018 paper in PNAS: “Molecular Mechanism of ATP versus GTP selectivity of adenylate kinase” The article is based on nice combo of NMR and X-ray!!! (Rogne, et al, PNAS, 2018, 115, 3012-3017 6F7U.pdb). 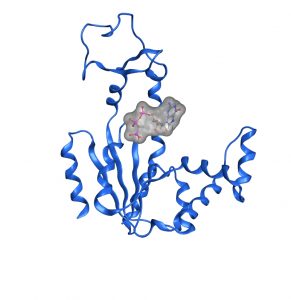 The basic paradigm in structural biology is that protein function is dictated by the three-dimensional structure. However, under cellular conditions, time dependent fluctuations of protein structures (dynamics) and temporal interactions with other macromolecules (such as proteins, nucleic acids and membranes) are indispensable for functionality. In my laboratory we investigate the relation between protein structure and dynamics to function. Our main experimental technique is high resolution protein NMR spectroscopy in solution. However, complementary techniques such as protein engineering, circular dichroism, isothermal titration calorimetry, molecular dynamics simulations and functional assays are integrated and essential in our research. We have two main research interests: (1) The role of enzyme dynamics in enzymatic catalysis, (2) Structure and dynamics of proteins that control infectivity in the bacterium Yersinia pseudotuberculosis.
The basic paradigm in structural biology is that protein function is dictated by the three-dimensional structure. However, under cellular conditions, time dependent fluctuations of protein structures (dynamics) and temporal interactions with other macromolecules (such as proteins, nucleic acids and membranes) are indispensable for functionality. In my laboratory we investigate the relation between protein structure and dynamics to function. Our main experimental technique is high resolution protein NMR spectroscopy in solution. However, complementary techniques such as protein engineering, circular dichroism, isothermal titration calorimetry, molecular dynamics simulations and functional assays are integrated and essential in our research. We have two main research interests: (1) The role of enzyme dynamics in enzymatic catalysis, (2) Structure and dynamics of proteins that control infectivity in the bacterium Yersinia pseudotuberculosis.
Research
Enzyme dynamics Structural flexibility is many cases intimately linked to the function of proteins and enzymes. Traditionally enzyme mechanisms have been deciphered by combining knowledge from static structures and kinetic measurements. In my lab we are interested in the time-dependent fluctuations of the enzyme and the coupling between fluctuations and function.One of the fundamental requirements for the viability of cellular organisms is that cellular chemical reactions occur on a time scale that is much faster than the turnover rate of global processes (such as cell division). This requirement is accomplished by the action of enzymes that are extraordinary biocatalysts accelerating chemical reactions. To accomplish their tasks enzymes have to perform several complex tasks; (i) binding of substrates weakly enough to avoid kinetic traps; (ii) extremely tight binding to the transition state compound; (iii) activation of functional groups on the substrate(s); (iv) optimal alignment of substrates to facilitate the chemical transformation into products and (v) dehydration of active sites. All these tasks are dependent on structural changes of the enzyme occurring on time-scales optimized for its biological function. In my lab we are interested in the time-dependent fluctuations of enzymes and the coupling between fluctuations and function. Adenylate kinase (AK) has emerged as one of the principal model systems to study the interplay between structure, dynamics and enzymatic activity. We have made several contributions to the understanding of conformational dynamics and activity by using the AK system. For example, we have recently shown that the magnitude of a pre-existing conformational equilibrium in AK modulates the catalytic parameters kcat and KM (JACS, 2012). We have also found that the mechanism of conformational change in response to ATP binding follow a “order-disorder-order” or “cracking”mechanism where specific segments unfold before refolding into the closed and active state (Nature Communications, 2010). In our PNAS, 2017 paper we managed to trap a high-energy AK state with subsequent characterization of its structure, dynamics and functional properties. The study revealed that substrate free AK dynamically samples the fully closed structure (which is the catalytically active state in presence of ATP and AMP). Now we are moving forward by addressing mechanisms of substrate selectivity and the driving forces for an induced fit transition.
Protein structure and dynamics in bacterial infections
Pathogenicity of the bacterium Yersinia pseudotuberculosis is dependent on translocation of effector proteins into eukaryotic host cells. The translocation process is accomplished with a multi-protein complex called the type III secretion system. Translocation is highly regulated and we study the structure and dynamics of proteins that are key regulatory players. One of our targets is YscU that is a membrane protein with a soluble cytosolic domain. Interestingly the soluble domain undergoes auto-proteolysis at a conserved NPTH motif. We have shown (PLoS ONE, 2012) that dissociation of the two folded polypeptides in YscU and secretion of the C-terminal polypeptide is crucial for effector protein secretion. We are continuing our reserach on YscU and also of a number of other proteins , all of which gives well-dispersed NMR spectra of high quality.
Instrumentation
Bruker 500 MHz spectrometer
Bruker 600 MHz spectrometer with cryoprobe
Bruker 850 MHz spectrometer with cryoprobe
Lab members
Dr. Per Rogne, Chief research engineer.per.rogne@chem.umu.se

Jonna Mattsson, Phd student. jonna.mattsson@umu.se
Chanrith Phouerk, Phd student. chanrith.phoeurk@umu.se
Léon Jona Schierholz, Phd student. leon.schierholz@umu.se
Sonja Tischlik, visiting PhD student from Konstanz University. sonja.tischlik@umu.se
Jhon Alexander Buitrago, Postdoc. rodriguez.jhon@umu.se
Tamás Milan Nagy, Postdoc. tamas.nagy@umu.se
ALUMNI
Ameeq Ul Mushtaq, postdoc Texas A & M University
Apoorv Verma, postdoc University of California Merceed
Michael Kovermann, Professor at University of Konstanz
Arun Gupta, postdoc at the University of Edinburgh
Jörgen Åden, research engineer at Umeå University
Oanh Ho, postdoc at Uppsala university
Stefan Frost, group leader at Roche Pharmaceuticals
Ulrika Olsson, Coordinator at the Karolinska Institute
Christoph Weise, NMR Spectroscopist at University of Evry Val d’Essonne
Contact Information
Magnus Wolf-Watz,
Professor
Department of Chemistry
Umeå University
SE-901 87
Sweden
magnus.wolf-watz@umu.se
Office phone: +46-90-786 7690