Structure and function of bacterial virulence factors
A bacterial biofilm is a complex network of bacteria and host tissue. We are using protein X-ray crystallography to study its molecular components in detail.
Our research is mainly directed towards understanding bacteria-bacteria and bacteria-host interactions, with the long term goal to shed light on biofilm formation and infection mechanisms. The first step towards understanding these molecular interactions is to study the structure and function of bacterial surface molecules that are involved. Investigating the structures of these surface proteins at an atomic level is the first step towards the possibility to interfere with certain interactions to hinder attachment and infection. The main technique that we are using is protein X-ray crystallography.
In addition, we are involved in several other projects where structure biology is one of the main techniques to answer biological questions.
Some of the projects:
1. Porphyromonas gingivalis proteins. P. gingivalis is a Gram-negative late colonizer that is the main cause of adult periodontitis. P. gingivalis known to hide from the human immune system and to be involved in the onset of systemic diseases such as cardiovascular disease and cancer. We are studying its surface molecules in order to understand how this bacterium can attach to the oral biofilm and to host cells. We have mostly focused on the Mfa1 fimbriae which belongs to fimbriae of type-V. Type-V fimbriae are only found in bacteroidetes bacteria.
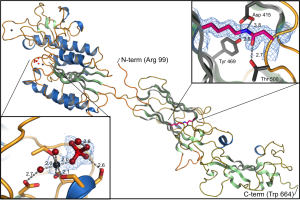
Mfa5, the tip protein if the P. gingivalis Mfa1 fimbriae. This is hitherto the only adhesin from a gram-negative bacteria that has been shown to be stabilized with an intramolecular isopeptide bond. The structure was solved by Thomas Heidler
2. Transportation of lipidated proteins. Type-V fimbrial proteins start as lipidated precursors attached to the inner membrane. Before they can be incorporated into the growing fimbria they have to be transported over the periplasm to the outer membrane. Gram-negative bacteria have several systems to transport proteins between the inner and outer membrane. The pathway that transports lipidated proteins is called the Lol-system. The proteins are lipidated at the inner membrane and recognized by the membrane protein complex LolCDE. The lipidated protein is next transferred to the soluble protein LolA that transfers it to the outer membrane where LolB receives it. We are studying the structure and function of LolA, LolB and the LolCDE complex. In addition we are investigating how LolA and LolB from differnt bacteria bind to antibiotics.
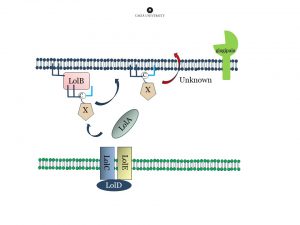
Fimbrial pre-proteins are lipidated at the inner membrane. Next they are transported over the periplasm via the LOL-stystem, and attached to the outer membrane. The protease “gingipain” is important for maturation of the protein (removal of the lipidated N-terminal extension) and subsequent polymerization.
3. Actinomyces oris proteins. A. oris is, as the oral streptococci, among the first colonizers of the oral biofilm. A. oris expresses two forms of pili, type-1 and type-2. We have solved the crystal structures of the main component the type-1 pili (FimP) as well as the enzyme, the sortase, that is needed to polymerize the subunits into mature pili. We are also interested in its disulfide forming mechanism, necessary to correctly fold pili proteins and other virulence factors.
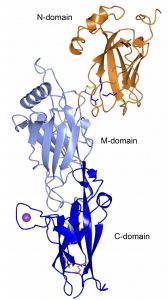
Crystal structure of FimP, the building block of the A. oris fimbria. The polymerization is catalyzed by a sortase. An adhesin, FimQ, is located on the tip of the fimbria. The structure is stabilized with intramolecular bonds and disulfide bonds
4. Cytotoxins. Vibrio cholerae produces a tripartite cytotoxin, MakABE that is secreted via the bacterial flagellum. We are studying the structure and function of these proteins. We have solved the crystal structures of the soluble forms of MakA, MakB and MakE using X-ray crystallography. The membrane bound forms will be studied using electron microscopy. In collaboration with Aftab Nadeem, Sun Nyunt Wai and Bernt Eric Uhlin (Umeå University).
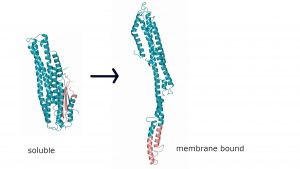
Crystal structure of MakA in its soluble form (left) and a model of the elongated membrane bound form (right). Together with MakB and MakE, that undergo a similar conformational change, a tripartite pore forming toxin is formed.
Group members:
Deepika Jaiman (PhD).
Previous group members:
Nina Forsgren (PhD 2010), Åsa Nylander (PhD 2013), Patrik Kloppsteck, (postdoc), Michael Hall (postdoc), Thomas Heidler (postdoc), Raghavendra Nagampalli (postdoc), Karin Ernits (postdoc) and Laura Modlich (master student).Sampath Yalamanchilli (postdoc), Nandita Bodra (postdoc).

